Using “Thinking about the Conceptual Foundations of the Biological Sciences” as a jumping off point. “Engineering biology for real?” by Derek Lowe (2018) is also relevant
Biological systems can be seen as conceptually simple, but mechanistically complex, with hidden features that make “fixing” them difficult.
Biological systems are evolving, bounded, non-equilibrium reaction systems. Based on their molecular details, it appears that all known organisms, both extinct or extant, are derived from a single last universal common ancestor, known as LUCA. LUCA lived ~4,000,000,000 years ago (give or take). While the steps leading to LUCA are hidden, and its precursors are essentially unknowable (much like the universe before the big bang), we can come to some general and unambiguous conclusions about LUCA itself [see Catchpole & Forterre, 2019]. First LUCA was cellular and complex, probably more complex that some modern organisms, certainly more complex than the simplest obligate intracellular parasite [Martinez-Cano et al., 2014]. Second, LUCA was a cell with a semi-permeable lipid bilayer membrane. Its boundary layer is semi-permeable because such a system needs to import energy and matter and export waste in order to keep from reaching equilibrium, since equilibrium = death with no possibility of resurrection. Finally, LUCA could produce offspring, through some version of a cell division process. The amazing conclusion is that every cell in your body (and every cell in every organism on the planet) has an uninterrupted connection to LUCA.
So what are the non-equilibrium reactions within LUCA and other organisms doing? building up (synthesizing) and degrading various molecules, including proteins, nucleic acids, lipids, carbohydrates and such – the components needed to maintain the membrane barrier while importing materials so that the cell can adapt, move, grow and divide. This non-equilibrium reaction network has been passed from parent to offspring cells, going back to LUCA. A new cell does not “start up” these reactions, they are running continuously through out the processes of growth and cell division. While fragile, these reaction systems have been running uninterruptedly for billions of years.
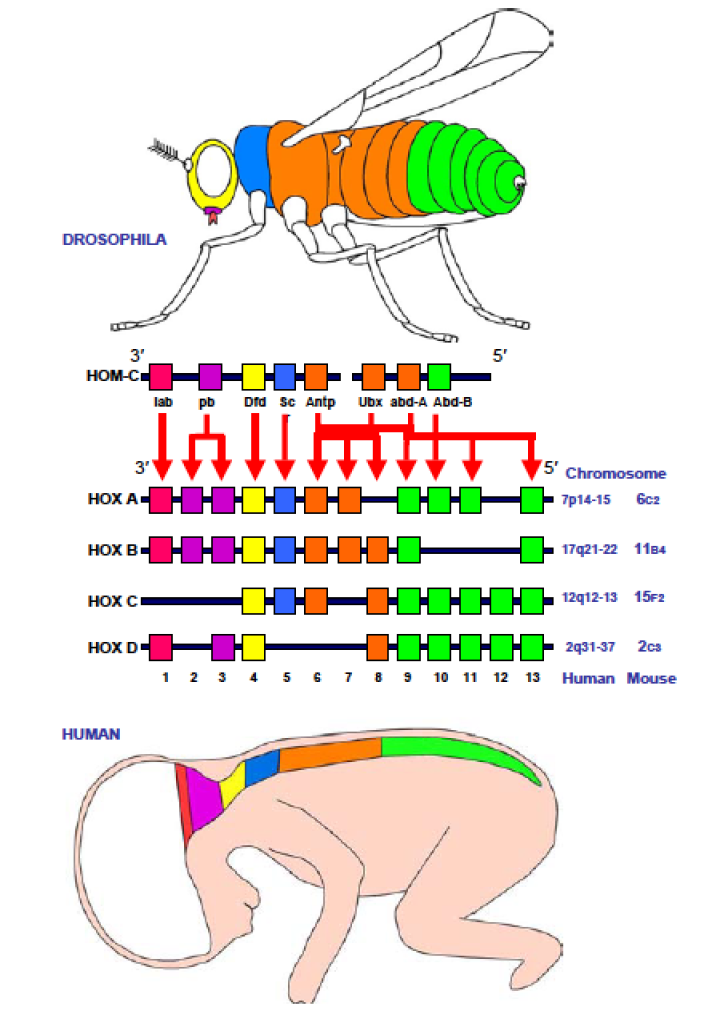
There is a second system, more or less fully formed, present in and inherited from LUCA, the DNA-based genetic information storage and retrieval system. The cell’s DNA (its genotype) encodes the “operating system” of the cell. The genotype interacts with and shapes the cell’s reaction systems to produce phenotypes, what the organism looks like and how it behaves, that is how it reacts to and interacts with the rest of the world. Because DNA is thermodynamically unstable, the information it contains, encoded in the sequences of nucleotides within it, and read out by the reaction systems, can be altered – it can change (mutate) in response to its environmental chemicals, radiation, and other processes, such as errors that occur when DNA is replicated. Once mutated, the change is stable, it becomes part of the genotype.
The mutability of DNA could be seen as a design flaw; you would not want the information in a computer file to be randomly altered over time or when copied. In living systems, however, the mutability of DNA is a feature – together with the effects of mutations on a cell’s reproductive success mutations lead to evolutionary change. Over time, they convert the noise of mutation into evolutionary adaptations and diversification of life.
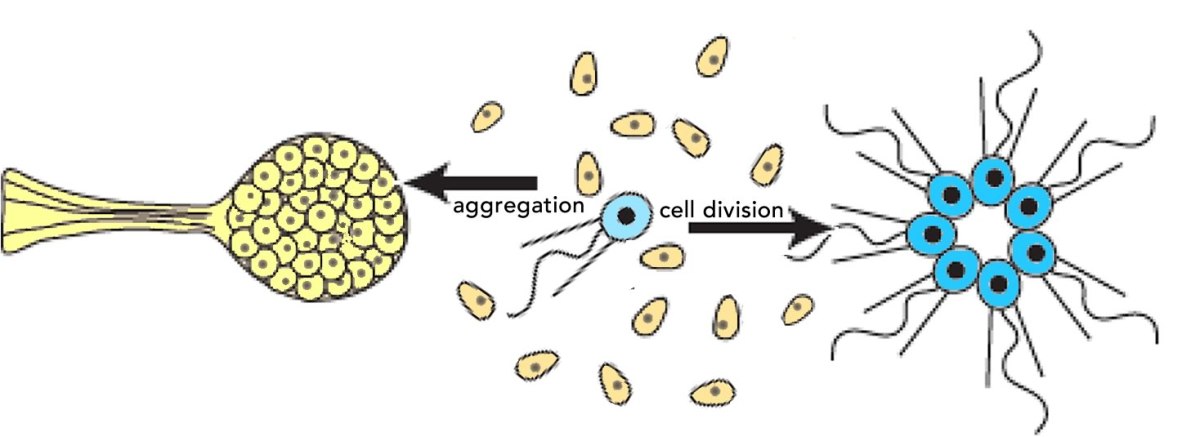
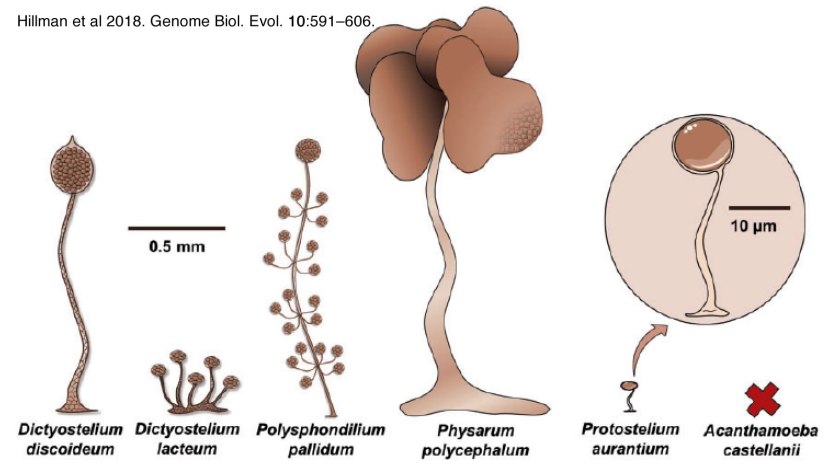
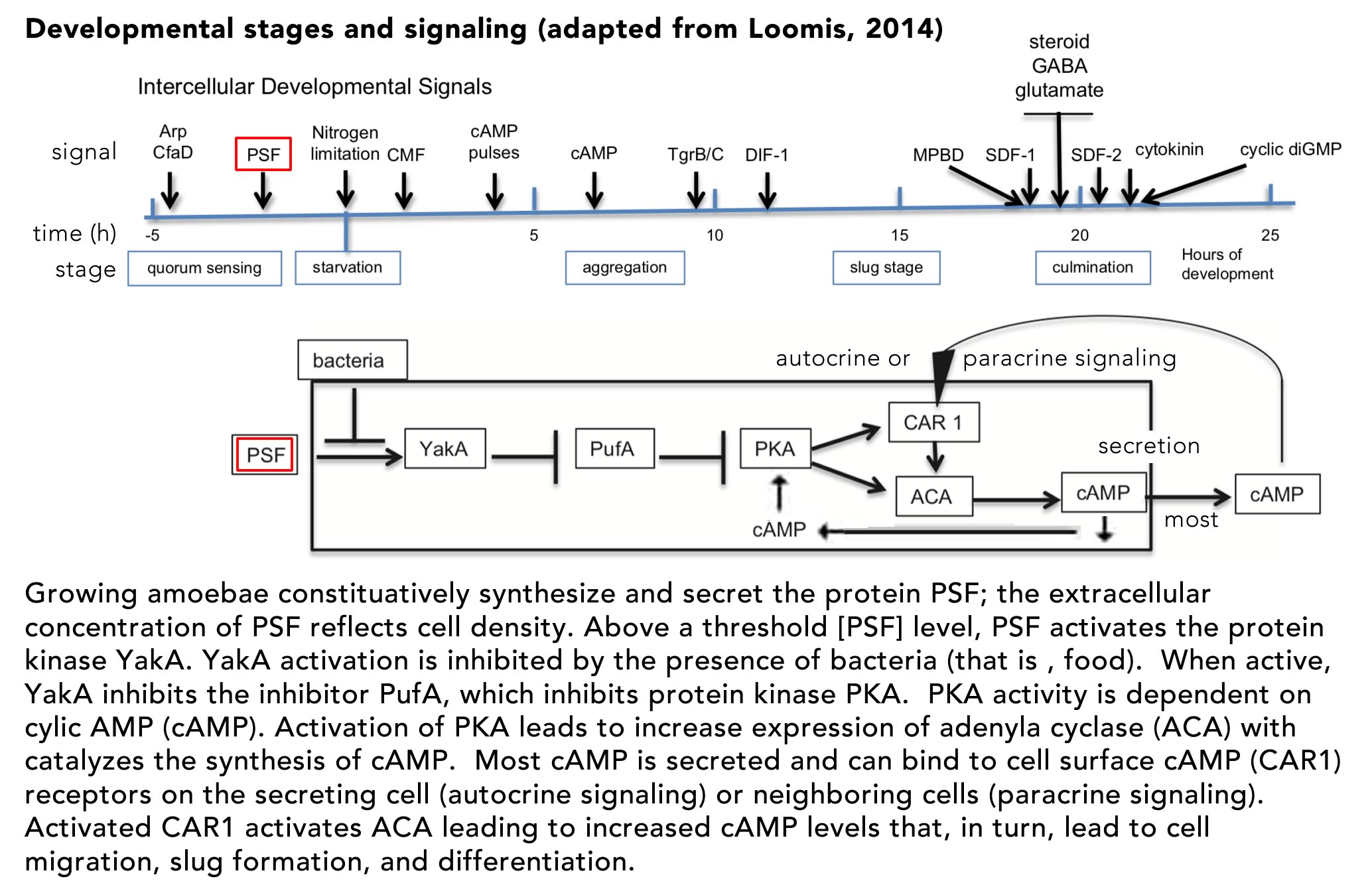
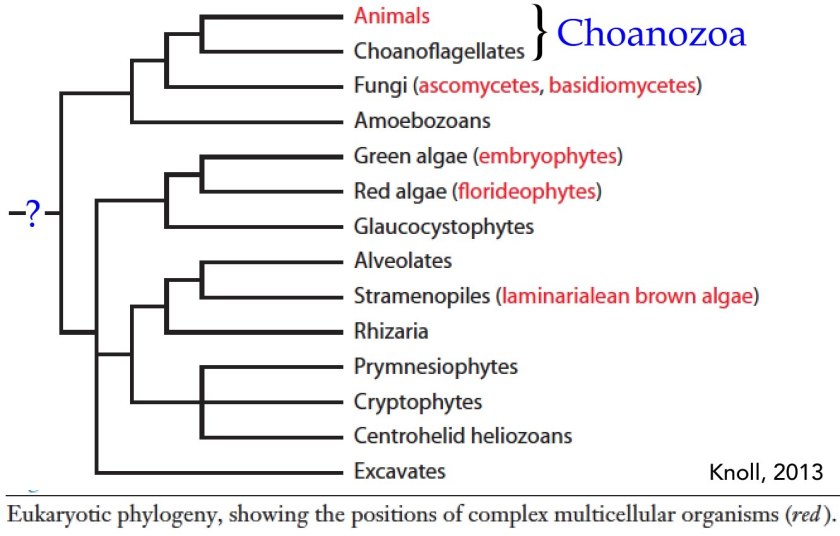
Organisms rarely exist in isolation. Our conceptual picture of LUCA is not complete until we include social interactions (background: aggregative and clonal metazoans). Cells (organisms) interact with one another in complex ways, whether as individuals within a microbial community, as cells within a multicellular organism, or in the context of predator-prey, host-pathogen and symbiotic interactions. These social processes drive a range of biological behaviors including what, at the individual cell level, can be seen as cooperative and self-sacrificing. The result is the production of even more complex biological structures, from microbial biofilms to pangolins and human beings, and complex societies. The breakdown of such interactions, whether in response to pathogens, environmental insult, mutations, politicians’ narcissistic behaviors and the madness of crowds, underlie a wide range of aberrant and pathogenic outcomes – after all cancer is based on the anti-social behavior of tumor cells.
The devil is in the details – from the conceptual to the practical: What a biologist/ bioengineer rapidly discovers when called upon to fix the effects of a mutation, defeat a pathogen, or repair a damaged organ is that biological systems are mechanistically more complex that originally thought, and are no means intelligently designed. There are a number of sources for this biological complexity. First, and most obviously, modern cells (as well as LUCA) are not intelligently designed systems – they are the product of evolutionary processes, through which noise is captured in useful forms. These systems emerge rather than are imposed (as is the case with humanly designed objects). Second, within the cell there is a high concentration of molecules that interact with one another, often in unexpected ways. As examples of molecular interactions that my lab has worked on, the protein β-catenin – originally identified as playing a role in cell adhesion and cytoskeletal organization, has a second role as a regulator of gene expression (link). The protein Chibby, a component of the basal body of cilia (a propeller-like molecular machine involved in moving fluids) has a second role as an inhibitor of β-catenin’s gene regulatory activity (link), while centrin-2. another basal body component, plays a role in the regulation of DNA repair and gene expression (link). These are interactions that have emerged during the process of evolution – they work, so they are retained.
More evidence as to the complexity of biological systems is illustrated by studies that examined the molecular targets of specific anti-cancer drugs (see Lowe 2019. Your Cancer Targets May Not Be Real). The authors of these studies used the CRISPR-Cas9 system to knock out the gene encoding a drugs’ purported target; they found that the drug continued to function (see Lin et al., 2019). At the same time, a related study raises a note of caution. Smits et al (2019) examined the effects of what were expected to be CRISPR-CAS9-induced “loss of function” mutations. They found expression of the (mutated) targeted gene, either by using alternative promoters (RNA synthesis start sites) or alternative translation start sites. The results were mutant polypeptides that retained some degree of wild type activity. Finally, in a system that bears some resemblance to the CRISPR system was found in mutations that induce what is known as non-sense mediated decay. A protection against the synthesis of aberrant (toxic) mutant polypeptides, one effect of non-sense mediated decay is to lead to the degradation of the mutant RNA. As described by Wilkinson (2019. Genetic paradox explained by nonsense) the resulting RNA fragments can be transported back into the nucleus where they interact with proteins involved in the regulation of gene expression, leading to the expression of genes related to the originally mutated gene. The expression of these related genes can modify the phenotype of the original mutation.

Biological systems are further complicated by the fact that the folding of polypeptides and the assembly of proteins (background: polypeptides and proteins) is mediated by a network of chaperone proteins, that act to facilitate correct, and suppress incorrect, folding, interactions, and assembly of proteins. This chaperone network helps explain the ability of cells to tolerate a range of genetic variations; they render cells more adaptive and “non-fragile”. Some chaperones are constitutively expressed and inherited when cells divide, the synthesis of others is induced in response to environmental stresses, such as increased temperatures (heat shock). The result is that, in some cases, the phenotypic effects of a mutation on a target protein may not be primarily due to the absence of the mutated protein, but rather to secondary effects, effects that can be significantly ameliorated by the expression of molecular chaperones (discussed in Klymkowsky. 2019 Filaments and phenotypes).
The expression of chaperones along with other genetics factors complicate our understanding of what a particular gene product does, or how variations (polymorphisms) in a gene can influence human health. This is one reason why genetic background effects are important when making conclusions as the health (or phenotypic) effects of inheriting a particular allele (Schrodi et al., 2014. Genetic-based prediction of disease traits: prediction is very difficult, especially about the future).
As one more, but certainly not the last, complexity, there is the phenomena by which “normal” cells interact with cells that are discordant with respect to some behavior (Di Gregorio et al 2016).1 These cells, termed “fit and unfit” and “winners and losers”, clearly socially inappropriate and unfortunate terms, interact in unexpected ways. The eccentricity of these cells can be due to various stochastic processes, including monoallelic expression (Chess, 2016), that lead to clones that behave differently (background: Biology education in the light of single cell/molecule studies). Akieda et al (2019) describe the presence of cells that respond inappropriately to a morphogen gradient during embryonic development. These eccentric cells are “out of step” with their neighbors are induced to die. Experimentally blocking their execution leads to defects in subsequent development. Similar competitive effects are described by Ellis et al (2019. Distinct modes of cell competition shape mammalian tissue morphogenesis). That said, not all eccentric behaviors lead to cell death. In some cases the effect is more like an ostracism, cells responding inappropriately migrate to a more hospitable region (Xiong et al., 2013).
All of which is to emphasize that while conceptually simple, biologically systems, and their responses to mutations and other pathogenic insults, are remarkably complex and unpredictable – a byproduct of the unintelligent evolutionary processes that produced them.
- Adapted from a F1000 review recommendation.